Research Summary
Our laboratory is interested in molecular mechanisms on RNA-mediated epigenetic regulations of gene expression and their role in Drosophila. The current research focuses: Epitranscriptome and its regulators, piRNA pathways, RNA-based immune system against transposable elements (TEs); histone modifications and their machinery; germ line development; high-throughput technologies; and genetic resource (NIG-FLY) for research in Drosophila.
Research Background
Transposable elements (TEs) occupy a large proportion of many eukaryotic genomes and their movement and accumulation play beneficial roles in the evolution of organisms. A new TE insertion at a genomic location can alter the transcriptome, which may compromise cell viability and threaten the integrity of the host genome. Thus, TEs should be recognized and repressed to maintain genome integrity. It is now over half a century since the discovery of TEs by Barbara McClintock, but large gaps still remain in our knowledge of exactly how TEs are recognized and repressed. The discoveries of PIWI-interacting RNAs (piRNAs) have given insights into the molecules that recognize TEs and how TEs are repressed.
The Argonaute (AGO) protein family functions in RNA interference (RNAi) in various organisms. 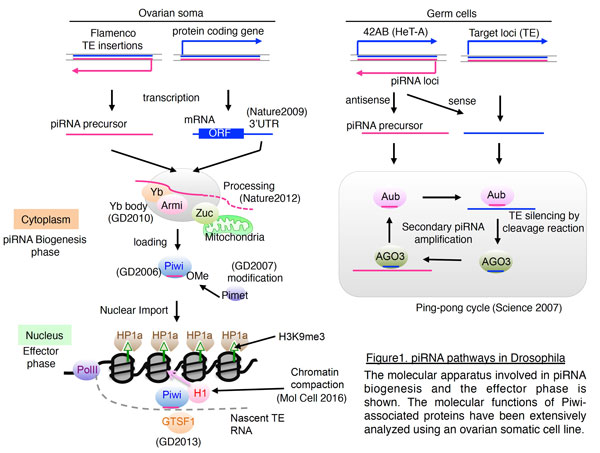 In Drosophila, there are five AGO genes. Of these, AGO1 and AGO2, classified as AGO clade members, function in microRNA (miRNA)- and small interfering RNA (siRNA)-mediated RNA silencing, respectively. The other three AGO genes, Piwi, Aubergine (Aub), and AGO3, are categorized as PIWI clade members. In 2002, our understanding of the PIWI clade genes was limited to their specific expression patterns and their involvement in germ cell development, but their molecular roles had not been determined. Generation of specific monoclonal antibodies against Piwi enabled us to identify a group of small RNAs, now called piRNAs that are complementary to TE mRNAs (Genes Dev 2006). From these findings, we proposed that Piwi utilizes small RNAs for their functions, similarly to the AGO clade proteins and plays a role in an innate immune system that selectively silences TEs in germ cells (Genes Dev 2006). Using fly genetics and biochemistry, we continued our investigation of the piRNA pathway and discovered that piRNAs are 2′-O-methylated at their 3′ ends and that a methyltransferase, named Pimet, is responsible for this modification (Genes Dev 2007). Moreover, our and Hannon’s groups demonstrated a distinct piRNA pathway, called the secondary pathway or Ping-Pong cycle, by which Aub and AGO3 carry out mutual cleavage (slicer) reactions and amplify the pool of piRNAs (Figure 1) (Science 2007; Cell 2007). Such amplified piRNAs are maternally deposited in the oocyte cytoplasm and transferred to progeny where specific TEs are silenced, according to inherited piRNA sequences (Brennecke et al. Science 2008).
In Drosophila, there are five AGO genes. Of these, AGO1 and AGO2, classified as AGO clade members, function in microRNA (miRNA)- and small interfering RNA (siRNA)-mediated RNA silencing, respectively. The other three AGO genes, Piwi, Aubergine (Aub), and AGO3, are categorized as PIWI clade members. In 2002, our understanding of the PIWI clade genes was limited to their specific expression patterns and their involvement in germ cell development, but their molecular roles had not been determined. Generation of specific monoclonal antibodies against Piwi enabled us to identify a group of small RNAs, now called piRNAs that are complementary to TE mRNAs (Genes Dev 2006). From these findings, we proposed that Piwi utilizes small RNAs for their functions, similarly to the AGO clade proteins and plays a role in an innate immune system that selectively silences TEs in germ cells (Genes Dev 2006). Using fly genetics and biochemistry, we continued our investigation of the piRNA pathway and discovered that piRNAs are 2′-O-methylated at their 3′ ends and that a methyltransferase, named Pimet, is responsible for this modification (Genes Dev 2007). Moreover, our and Hannon’s groups demonstrated a distinct piRNA pathway, called the secondary pathway or Ping-Pong cycle, by which Aub and AGO3 carry out mutual cleavage (slicer) reactions and amplify the pool of piRNAs (Figure 1) (Science 2007; Cell 2007). Such amplified piRNAs are maternally deposited in the oocyte cytoplasm and transferred to progeny where specific TEs are silenced, according to inherited piRNA sequences (Brennecke et al. Science 2008).
Discovery of distinct piRNA pathways led us to question how piRNAs are produced. To address this issue, we developed an ovarian somatic cell-line, OSC, and conducted RNAi analyses to identify the responsible factors and uncover the mechanism of piRNA biogenesis (Nature 2009; Genes Dev 2010). 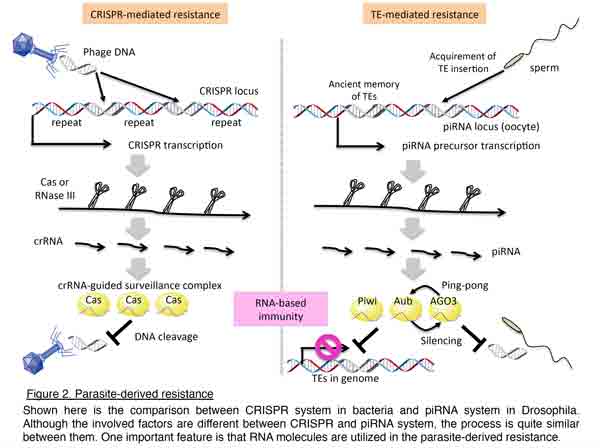 We have described roles for the cytoplasmic Yb body components, Armitage (Armi) and fs(1)Yb, in somatic piRNA biogenesis and proposed that a functional Piwi–piRNA complex is formed and inspected in Yb bodies before its entry into the nucleus to exert TE silencing. In collaboration with Dr. Nureki, we have also demonstrated that Zucchini (Zuc) acts as an endoribonuclease essential for piRNA biogenesis (Nature 2012). These findings together indicate that piRNAs are selectively produced from the genome, and are recognized and processed in a cooperative manner by dozens of protein factors. From the stepwise biogenesis and the repressive functions of piRNAs against invasive TEs, piRNA-mediated genome defense is recently considered similar to the CRISPR system in bacteria (Figure 2).
We have described roles for the cytoplasmic Yb body components, Armitage (Armi) and fs(1)Yb, in somatic piRNA biogenesis and proposed that a functional Piwi–piRNA complex is formed and inspected in Yb bodies before its entry into the nucleus to exert TE silencing. In collaboration with Dr. Nureki, we have also demonstrated that Zucchini (Zuc) acts as an endoribonuclease essential for piRNA biogenesis (Nature 2012). These findings together indicate that piRNAs are selectively produced from the genome, and are recognized and processed in a cooperative manner by dozens of protein factors. From the stepwise biogenesis and the repressive functions of piRNAs against invasive TEs, piRNA-mediated genome defense is recently considered similar to the CRISPR system in bacteria (Figure 2).
We have also made important progress in transcriptional silencing by Piwi-piRNA complex. In contrast to Aub and AGO3, Piwi does not require cleavage of TE transcripts (Genes Dev 2010), but involves chromatin modification of TE loci (Sienski et al. Cell 2012). A candidate gene approach in OSCs allowed us to identify DmGTSF1, a nuclear protein that interacts with Piwi. Females carrying a P-element insertion in DmGTSF1 showed sterility and possessed severely deformed ovaries, similar to those observed in piwi mutants. ChIP analyses revealed strong enrichment of RNA polymerase II and less efficient enrichment of H3K9me3 at genomic TE loci, indicating that DmGTSF1 is an integral factor in Piwi–piRISC-mediated transcriptional silencing (Genes Dev 2013). In addition, we recently demonstrated that Piwi physically interacts with linker histone H1 and regulates the interaction of multiple chromatin components, including H3K9me3 and heterochromatin protein 1a (HP1a), with target loci to maintain silencing of TEs (Mol. Cell 2016).
Research Projects
Studies of piRNAs have stimulated a broad interest in epigenetics and TE biology. Currently, we are interested in:
(1) How does Piwi-piRNA complex induce and establish a silent chromatin state?
(2) How do animals defend their genomes against the enormous complexity of TEs in germ and somatic cells?
(3) To what degree, and by what mechanisms, do multiple epigenetic pathways contribute to germ cell development in Drosophila?
To answer the above questions, we are conducting approaches include culture cell assays, biochemical reconstitutions, cell imaging techniques, molecular experiments, bioinformatics and genetic experiments. To our advantage, we also preserve and accumulate Drosophila strains and distribute them all over the world through NIG-Fly. So, we are also welcome your proposals for new genetic resources accelerating fly research. Notably, we are currently conducting some projects that are not described here. If you are interested in our labratory, please feel free to ask me (K. Saito).